Transform-based Evolvable Features
Here we approach the problem of automatically finding means of classifying an image database. That is, if one has a labelled database of images, how can one develop a system which will correctly classify them and new instances? For instance, given a number of images of cells, we might know which ones are healthy and sick. Can we automatically generate a program which will distinguish between healthy and sick cells presented in the future?
Our approach to image classification is to use a transformation stage in the processing of an image. That is, via evolutionary computation, we attempt to extract a mathematical description of the important sub-patterns in an image, and then use those mathematical descriptions to transform the database into a more recognizable version.
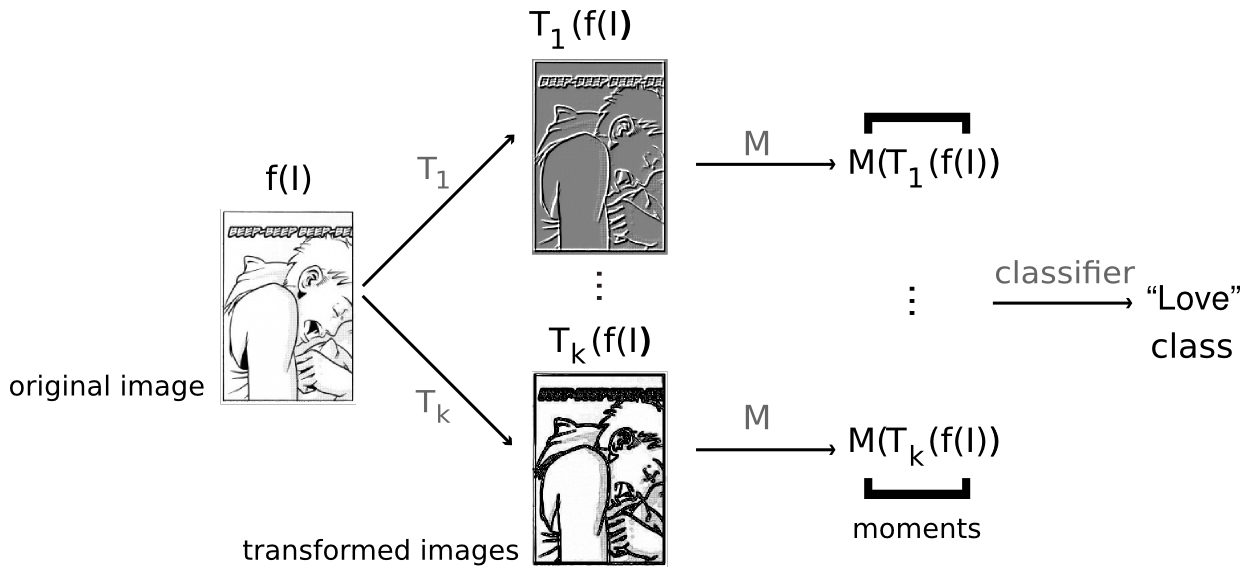
An illustration of the process of applying a TEF to an image.
Given some image, a classifier, and a collection of transforms, we can classify the image as follows:
- We apply the k transforms to the original image, producing k transformed images.
- We extract m numerical values from each transformed image, using a standard dimensionality reducing technique (usually statistical moments). This produces a vector of mk values
- That vector is fed to a classifier to determine the class
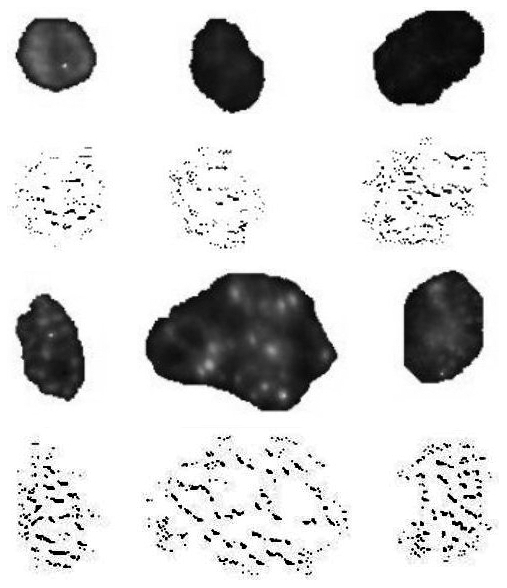
Action of one of the transforms on cell images. Rows 1 and 3 show original cell images, healthy cells at the top and sick cells on the bottom. Rows 2 and 4 show transformed versions.
This same process is used to find the transforms. Transforms are represented as genetic programming graphs. Initially, they are generated randomly (and thus are not very good). But, using our target classifier as a wrapper, we can gradually evolve better transforms, moving towards transforms that are adept at extracting sub-patterns that help classify the image database.
The use of an evolutionary system optimizing performance under a wrapper approach has some draw-backs: firstly, the system is an offline-learner only; secondly, transforms tend to be classifier-specific; and thirdly, as with many GP systems, overfitting is a major concern. The first two problems we simply accept, but the third, over-fitting, is a major concern. We combat over-fitting through several means: by limiting the choice of dimensionality reducing moments, or by using a form of competitive co-evolution.
We have shown that in some contexts, an appropriate solution size (number of transforms, size of pixel neighbourhood) can be determined automatically by evolution. As such, we believe that TEFs are an appropriate approach when the relevant patterns in a database are poorly understood or unknown.
TEFs have thus far been applied to medical cell classification (the recognition of cells with a form of muscular dystrophy, OPMD CEC 2009, GECCO 2012), and to the recognition of artistic styles (using the CAPD database, CEC 2010). In the case of OPMD, we matched the state-of-the-art.
We also applied TEFs to hand-written character recognition, where we obtained performance better than a naive techique (i.e. direct application of classifiers from Weka), but approximately 5% worse than state-of-the-art. (Alas, this work is unpublished). We attribute this less-than-ideal performance to the fact that writing systems have been designed to be optimal for human visual recongition, and thus, traditional features based on human vision perform very well.
In the case of recognition of artistic style, we managed to go further, and use the generated transforms as a basis for a distance metric. That is, we managed to create a measure of distance from an arbitrary image to an artist's style. The measure was able to rank an artist's examples as closer to his or her style relative to random images drawn from a search engine. Further, some qualifying phenomena were discovered: the measure ranked Manga images as close to each other, but far from other artists' styles; it found an image by one of the CAPD artists from a different project in the control group; it recommended similar artists from the internet whose styles, IOHO, were similar to the artists in question. See our CEC 2010 paper for details.